Yuhong Zuo
Structural Studies of Bacterial Transcription Initiation and Regulation
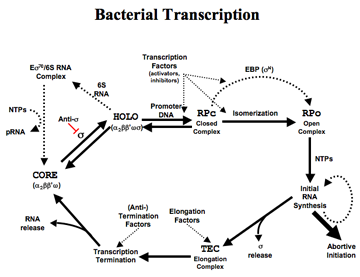
My research at Yale focuses on bacterial RNA polymerase, the cellular RNA synthesizing machinery, more specifically, on bacterial transcription initiation and regulation. RNA polymerase is one of the largest protein complexes in the cell and is responsible for transcribing RNA using DNA as templates. Significant progresses have been made on the bacterial transcription elongation in recent years, but the details of the bacterial transcription initiation remain elusive, largely due to the highly dynamic properties of the initiation process. The goal of this project is to capture the steps in transcription initiation by X-ray crystallography.
Structural and Mechanistic Studies of Exoribonucleases
My previous work at University of Miami focused on RNA degrading enzymes, quite an opposite to my current research focus. Specifically I was trying to understand the mechanisms that define the various substrate specificities and biological functions of exoribonucleases, enzymes cleave RNA molecules from their ends, on a structural basis. This effort led to high resolution stuctures of several Escherichia coliexoribonucleases and great details of their mechanisms of actions.
Selected Publications
Zuo, Y., Vincent, H.A., Zhang, J., Wang, Y., Deutscher M.P. & Malhotra, A. (2006). Structural basis for processivity and single-strand specificity of RNase II. Mol. Cell 24, 149-156.
Zuo, Y., Wang, Y. & Malhotra, A. (2005). Crystal structure ofEscherichia coli RNase D, an exoribonuclease involved in structured RNA processing. Structure 13, 973-984.
Zuo, Y. & Deutscher, M.P. (2002). The physiological role of RNase T can be explained by its unusual substrate specificity.J. Biol. Chem. 277, 29654-29661.
Zuo, Y. & Deutscher, M.P. (2001). Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acid Res. 29, 1017-1026.
